In Silico Prediction of SSRs and Functional Annotation of ESTs from Catharanthus Roseus
Pravej Alam*, Thamer Albalwai
Department of Biology, College of Science and Humanities, Prince Sattam bin Abdulaziz University, Al-Kharj 11942, Saudi Arabia
*Email: alamprez @ gmail.com
ABSTRACT
Catharanthus roseus (periwinkles) belongs to the Apocynaceae family with great anti-cancer, anti-diabetic, and hepatoprotective values. Due to the large number of active molecules accumulated in these plants, they are of particular concern, especially in the pharmaceutical sector. The availability of ESTs gave the genetic algorithm of the plant to differentiate between the species accessions at the genetic level. The high-throughput method used for mining and detection of microsatellites (SSRs) embedded in ESTs gave a new insight for molecular markers’ development. 19899 ESTs were retrieved, examined by NCBI EST dB and assembled 2692 to get full-length contigs sequences. 338 microsatellites (SSR) loci were predicted with an average of SSR per 9.33 kb of EST though MISA-web tools out of 2692 contigs. Furthermore, trinucleotide, a well-known SSR was examined and found to be the most favorable repeats' type (26.62%) followed by dinucleotide (24.22), mononucleotide (48.22%), and hexanucleotide (0.3%) types. The highest frequency of (A/T)n was reported in this finding followed by (AAG)n. The simple sequence repeats (SSR) extracted from C. roseus EST's data were used as molecular tools for genetic characterization in the present study. These predicted SSRs can be significantly used for constructing the genetic maps and also for differentiating the accession between the species.
Key words: C. roseus, ESTs, Contigs, SSR.
INTRODUCTION
Madagascar periwinkle (Catharanthus roseus L.) is a dicotyledonous and miracle medicinal plant with antitumor bioactive compounds [1-3]. The plant is domesticated and cultivated worldwide for ornamental and medicinal use [4, 5]. C. roseus is also one of the best sources for terpenoid indole alkaloids synthesized with a wide range of plant metabolites [6-8]. It has antidiabetic properties when the alcoholic extract of the plant is given to the streptozotocin-induced diabetic rats and shows a remarkable effect in lowering of glycemia in both diabetic and normal rats [9, 10]. Since many species have been evolved worldwide, it is difficult to segregate the plant at the species level.
Molecular markers (RAPD, AFLP, and SSR) are extensively used for analyses of genetic diversity, mapping of loci or gene, and marker-assisted selection in breeding technology [11-13]. These microsatellites or SSRs are distributed in plant genomes showing co-dominance and reproducibility. It has been frequently used for genetic scrutiny in breeding technology rand it is recognized as a consistent and crucial method in plant genetics [14]. The advantages of SSRs development gave efficient hypervariability, codominant, and widespread genomic to differentiate the plant species at an inclusive spectrum [15].
METHODOLOGY
19899 C. roseus express sequence tags (ESTs) were retrieved from dB EST of NCBI for the analyses of SSRs. The EST sequences were passed through sequence cleaning and masking, through high throughput web application EGassembler [16] contrary to the NCBI Vec screen (https://www.ncbi.nlm.nih.gov/tools/vecscreen/) to remove the vector contaminant, poly A/T, and short sequences (adapter). The following parameters' minimum match (>10) and minimum score (20) with no stretch of (A/T)n the existing step-wise process were applied in EGassembler.
Finally, in a non-redundant dataset of 2692 contigs, CAP3 program (EGassembler) was used to assemble the contigs for further analyses. MISA web (https://webblast.ipk-gatersleben.de), a web SSR finder developed by Beier et al. [17] was used to predict the EST-SSR loci.
Functional Annotations
The functional annotations with references to the biological process of assembled EST-contigs (2692) were predicted by the BLAST2go program obtained from Omics data. Based on NCBI BLAST and gene ontology, the biological function was predicted.
RESULTS AND DISCUSSION
A total of 19899 redundant ESTs’ sequences were carried out for SSR analyses, retrieved from NCBI EST dB having 10196333 bp in C. roseus genome. During pre-processing, the sequence clean, masking of vector contaminant, low complexity sequences, and Poly A/T tails were examined and assembled effectively from 100087bp C. roseus ESTs. After mining of 19899 ESTs (5963 trimmed), 2692 contigs were finally generated to obtain the hypervariable class, I microsatellites i.e SSRs. The MISA web application (https://webblast.ipk-gatersleben.de) was used to screen the SSR though the mining program to search the 1-6 nucleotide repeat motifs. In this program, it was observed that only 338 hypervariable SSR loci were developed from 2692 contigs (Table 1). The SSR loci frequency was 9.33 kb per ESTs analyzed in C. roseus.
Table 1: The results of microsatellite search of Catharanthus roseus ESTs
|
Parameters |
Values |
|
Total number of ESTs |
19899 |
|
Total sequence analyzed in bp |
10196333 |
|
Total masking in bp |
100087 |
|
EST after vector and Poly A/T removal |
19868 |
|
Total number of singletons |
5646 |
|
Total number of sequences examined: |
2692 |
|
Total size of examined sequences (bp): |
1911399 |
|
Total number of SSR loci located |
338 |
|
Frequency of SSR loci in Catharanthus roseus EST |
1 per 9.66 kb |
The obtained frequency per SSR loci from C. roseus contigs was found satisfactory and in agreement with other findings in many plants (Figure 1). It is also noted that this finding is also in agreement with previous studies reported in rice (3.4), soybean (7.4), and maize (8.1) [18-20].
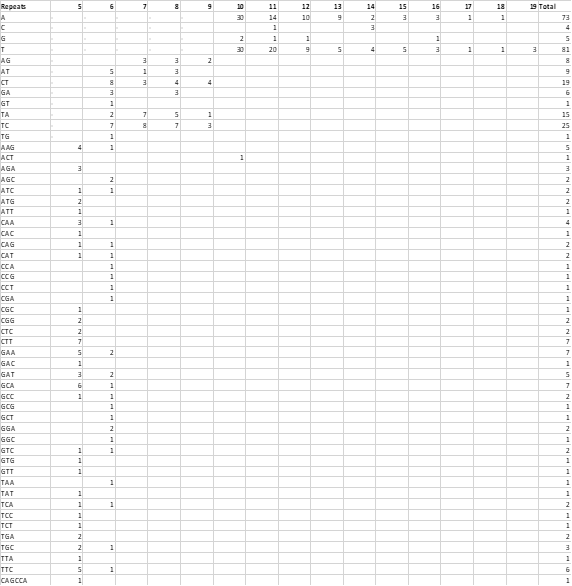
Figure 1 : Frequency of identified SSR motifs from assembled ESTs of C. roseus
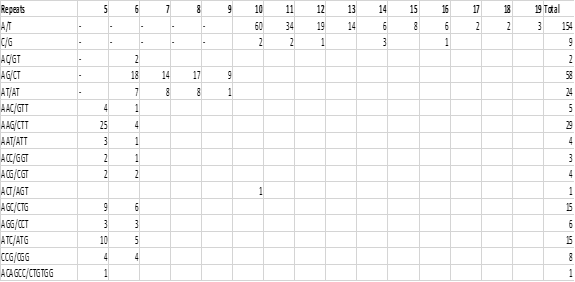
Figure 2 : Frequency of classified repeat types (considering sequence complementary) from assembled ESTs of C. roseus
The developed microsatellites or SSRs were characterized as the signature sequences in terms of simple motif type or compound motif or both. The obtained 338 SSRs from C. roseus are simple repeat motifs consisted of mononucleotide to hexanucleotide. The maximum frequency of motif was recorded as mononucleotide (48.22%) trailed by the trinucleotide (26.62%), dinucleotide (24.22), and hexanucleotide types (0.29%). The finding of trinucleotide repeat as the main single sequence repeats ; played an important role in previously reported plants (Figure 2) [21, 22]. The trinucleotide was earlier reported in wheat (32%), and sorghum (49%) with the signature sequence (CCG)n. similarly, the (AAG)n motif repeats have shown the maximum or rich motif in the trinucleotide repeat of Gossypium barbadense and Curcuma longa, which is in agreement with our findings (AAG)n) as shown in Table. 4 [23, 24]. The obtained dinucleotide (non-trimeric repeats) rather than trinucleotide SSRs may not be qualified as the best SSRs nomenclature due to some mutations [20]. The resulted SSR repeat motifs obtained from the ESTs-contigs were shown in Figure 3 as A/T (45.56%), AG/CT (17.16%), AAG/CTT (8.58%), AGC/CTG (4.44%), ATC/ATG (4.44%), CCG/CGG (2.37), and AGG/CTT (1.78%) rich frequency respectively. The furthermost common repeat motifs were predicted as A/T, AG/CT, AAG/CTT, ACAGCC/CTGTGG with the frequency of 45.56%, 17.16%, 8.58%, 31.42%, 15.68% and 8.98% and 0.3%, respectively (Table 2). The biological function was also predicted by using the method of the Blast2Go program on the Omics box and they were recorded as GO:0008150 (biological process), GO:0050896 response to stimulus and GO:0051716 (cellular process) followed by others [25, 26] (Figure 4; Table 3).
Table 2: Maximum frequency of SSR repeats based on nucleotide repeats from assembled ESTs of C. roseus
|
Repeats based on nucleotide |
SSRs repeats |
% Frequency |
Profuse motif |
% Frequency |
|
Mononucleotide |
163 |
48.22 |
A/T |
45.56 |
|
dinucleotide |
84 |
24.22 |
AG/CT |
17.16 |
|
Trinucleotide |
90 |
26.62 |
AT/AT |
7.10 |
|
Hexanucleotide |
1 |
0.29 |
AAG/CTT |
8.58 |
|
Total |
338 |
- |
AGC/CTG |
4.44 |
|
- |
- |
- |
ATC/ATG |
4.44 |
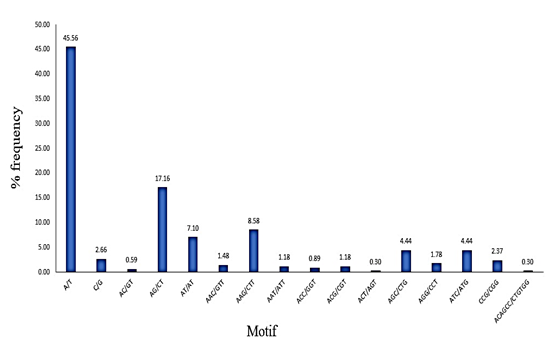
Figure 3: Graphical representation of SSR-motif with their distribution frequency analyzed from assembled EST of C. roseus
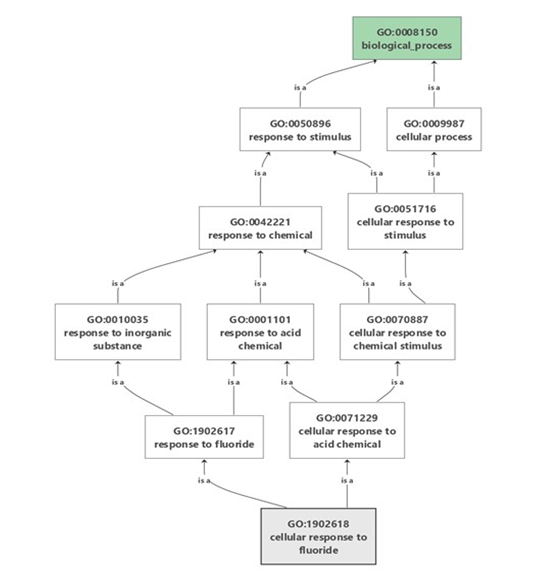
Figure 4: Gene ontology-based functional annotation of EST-contigs obtained from C. roseus showing the biological functions
Table 3: Functional annotation of ESTs-contigs for biological process
|
Level |
GO ID |
GO Name |
GO Type |
Parents (ACC) |
Parents (Name) |
|
1 |
GO:0008150 |
Biological process |
Biological Process |
- |
- |
|
2 |
GO:0050896 |
Response to stimulus |
Biological Process |
GO:0008150 |
Biological process |
|
2 |
GO:0009987 |
Cellular process |
Biological Process |
GO:0008150 |
Biological process |
|
3 |
GO:0051716 |
Cellular response to stimulus |
Biological Process |
GO:0009987, GO:0050896 |
Cellular process, response to stimulus |
|
3 |
GO:0042221 |
Response to chemical |
Biological Process |
GO:0050896 |
Response to stimulus |
|
4 |
GO:0010035 |
Response to inorganic substance |
Biological Process |
GO:0042221 |
Response to chemical |
|
4 |
GO:0001101 |
Response to acid chemical |
Biological Process |
GO:0042221 |
Response to chemical |
|
4 |
GO:0070887 |
Cellular response to chemical stimulus |
Biological Process |
GO:0051716, GO:0042221 |
Cellular response to stimulus, response to chemical |
|
5 |
GO:0071229 |
Cellular response to acid chemical |
Biological Process |
GO:0001101, GO:0070887 |
Response to acid chemical, cellular response to chemical stimulus |
|
5 |
GO:1902617 |
Response to fluoride |
Biological Process |
GO:0010035, GO:0001101 |
Response to inorganic substance, response to acid chemical |
|
6 |
GO:1902618 |
Cellular response to fluoride |
Biological Process |
GO:0071229, GO:1902617 |
Cellular response to acid chemical, response to fluoride |
CONCLUSION
338 non-redundant hypervariable SSR type I obtained from EST of C. roseus using SSR predicted from MISA web tool is found to be reproducible, cost-effective, and time-saving. These 338 non-redundant SSR information may give a better platform to understand the genetic variation and help in the genome mapping of C. roseus. The functional annotation also provided information about ESTs involved in various processes. It may be used to provide the deep functional gene network information that is involved in particular metabolite synthesis with their deep functional annotations in C. roseus genome.
ACKNOWLEDGMENT
This Publication was supported by the Deanship of Scientific Research at Prince Sattam bin Abdulaziz University.
REFERENCES