Isolation, Characterization and Molecular Identification of Oil Degrading Streptomyces sp.MB9 From Soils
Mona Othman I. Albureikan
Biology Department, Faculty of Science, King Abdulaziz University, Saudi Arabia.
Email: mona.albureikan @ gmail.com
ABSTRACT
Uses of oil products increased each year which lead to soil and water contamination and many environmental problems. Bioremediation or speed up the degradation processes of these contaminants from soil and water is considered an efficient strategy for treatment. Fifteen actinomycete isolates were obtained from four oil-contaminated soil samples. All isolates were grown in liquid medium containing crude oil as carbon source. Five actinomycete isolates showed the highest growth, thus they were screened for Biosurfactant productions by oil spreading, surface tension, and hemolytic assay. The most active isolate was identified using morphological and molecular identification as Streptomyces sp. MB9. The identified Streptomyces sp. MB9 was used for biodegradation of different concentrations of hydrocarbons for 10 days and growth and percentage of degradation were determined. Maximum degradation (100%) was for fluorene (500 ppm) and Naphthalene (500 ppm) after 5 days of incubation while lower degradation rates were for Phenanthrene, Anthracene, and Pyrene. In conclusion, the selected Streptomyces sp MB9 played an important role in oil degradation.
Key words: Biodegradation, aromatic hydrocarbons, Streptomyces sp., actinomycetes, Biosurfactant, oil.
INTRODUCTION
Petrol composed of a complex mixture of different heavy polycyclic aromatic hydrocarbons in addition to other constituents which led to chronic and harmful hazards to microbes in soil and water, human, and animals including carcinogenicity and mutagenicity for human and animal cells [1-5]. One of the most dangerous environmental problems during the 21 century is oil-contaminated soil and water [6-8] which affect aquatic life, plant metabolism, and growth of bacteria and fungi [9-11]. Clearing of soil from oil by chemical or physical techniques is difficult or uneffective [9] while bioremediation techniques are more effective [12, 13]. Using bacteria in biodegradation processes is of great interest where many isolates from oil-contaminated soils, are utilizing crude oil as a carbon source. Pseudomonas earuginosa, Flavobacterium sp., Bacillus sp. and Acinetobacterium calcoaceticum, under laboratory conditions singly or a consortium of these isolates, are capable of utilizing oil and there is a clear relationship between bacterial growth and degradation rate [14, 15] Nocardia, Alcanivorax, Mycobacterium, Bacillus sp., and Pseudomonas were used effectively for oil removal from the environment [16-18]. Modified diesel engine oil medium was used to isolate Micrococcus sp. and Pseudomonas sp.and the two isolates hydrolyze hydrocarbon after 25 days of incubation with a degradation rate of 67.57 % for Pseudomonas sp., 52.95 % for Micrococcus sp. and 89.98 % for the mixture of the two bacteria [19]. The cyanobacterium strain AL-12 used 98% of alkanes in motor oil after 50 days and normal alkane (n-alkane) can be biodegraded by Rhodococcus sp. and Gordonia sp. [20-22]. Different bacterial genera used crude oil under optimized conditions as carbon sources[23, 24] while Obayoriet al. [25] found out that Pseudomonas aeruginosa from contaminated soil was the highest petroleum hydrocarbon degrader. Moreover, Pseudomonas putrefacient, Klebsiella pneumonia, Pseudomonas alcaligenes, Klebsiella aerogenes, and Bacillus coagulans degraded oil by 68%, 62%, 59%, 58% and 45%, respectively [26]. Similarly, the rate of degradation was 47 - 66 % of aliphatic compounds in the engine oil, after 60 days of incubation [1]. The presence of hydrocarbon-degrading genes like catechol 2, 3 dioxygenase gene and alkane monooxygenase was confirmed by many authors [13, 27, 28]. In natural environments, oil biodegradation is habitually limited due to substrate low availability for the microorganisms and low nutrients contents, thus improving the efficiency of degradation in both soil and water is important. Increasing the solubility of oil by biosurfactants, which are themselves completely biodegradable can significantly enhance the biodegradation process [29]. Bacteria during their growth produced a natural surfactant called Biosurfactant which attracted much attention in the different industries due to high biodegradation, less toxicity and high performance under extreme conditions [30-32]. The lipopeptide, surfactin is the most effective and powerful biosurfactant obtained from the bacteria of genus Bacillus sp.[33-35] and during fermentation, the surface tension was decreased from 72 to 27 mN/m [30]. This study aimed to isolate and characterize oil-degrading bacteria from local contaminated soil.
MATERIALS AD METHODS
Chemicals used in this study
Analytical and high pure grade hydrocarbons, Naphthalene, phenanthrene, anthracene, pyrene, and Fluorene were purchased from Sigma-Aldrich while all used agar medium were from Himedia, India. The oil sample was collected from Armco, Saudi Arabia. Stock solutions of each hydrocarbon in n-hexane (Fluka) were prepared.
Sample collections and analysis
Top samples of four oil-contaminated soils at a depth 1-15 cm were collected in a sterile plastic bag from the oil service station, Jeddah, Saudi Arabia and immediately transferred to the lab. for analysis and bacteria isolation. The soil was air-dried and pH value of soil was determined in soil suspension (1:1 w/v) using pH meter according to MAFF [36] while total soil organic carbon and EC were determined according to McCauley et al. [37].
Total hydrocarbon content of the soil
The total hydrocarbon content of each soil sample was determined after extraction of soil (100 g) with methanol in the presence of KOH (3 g) under relaxation. The reflex mixture was filtrated, re-extracted with 25 ml hexane, cleared using a silica gel column, and the clean extract was dried and weighted [38, 39].
Isolation of actinomycete isolates
About 15 actinomycete isolates were obtained on Starch nitrate agar medium from the collected soil samples, purified and preserved on the same medium [40].
Rapid Screening for oil-degrading bacteria
The obtained actinomycete isolates were screened for their ability to degrade crude oil in Minimal salt broth (MSB) medium composed of g/l: 2.5 NH4Cl, 5.46 KH2PO4, 4.76 Na2HPO4, 0.2 MgSO4, 5.0 NaCl, 5g/l crude oil as a sole carbon source (pH 7.4). Each flask was inoculated with 2 ml of broth culture (8x107 cfu/ml) and all flasks were incubated at 45°C for 10 days. About 0.5 ml of the crude culture was used to inoculate agar plats of the same medium and crude oil ethereal solution (10% w/v) was sprayed over the agar plate surface. After solvent vaporization, a thin layer of oil remained on the agar surface. After incubation at 45°C for 5 days, the organism that formed a clear zone around the colony was considered as crude oil degraders and selected for detail studies [41]. Degradation of different hydrocarbons was confirmed using the same medium.
Screening biosurfactantproduction
All isolates were grown in Basal Broth medium at pH 7 (7% olive oil (v/v), 1% dextrose (w/v) 3% peptone (w/v), for 10 days at 45°C and 150 rpm. Cells were collected after centrifugation at 3,000 rpm for 15 min and the supernatant was used for screening for biosurfactant production, Oil spreading assay, Hemolytic assay, Drop collapse test, Emulsification test, and Surface tension assay.
Morphological, physiological and biochemical characterization
The best oil-degrading actinomycete was characterized and identified. Growth on different agar media [42], bacterial morphology, carbon, and nitrogen utilization and sensitivity to some antibiotics were detected [37]. Diaminopimelic acid, whole-cell sugar, Phospholipid types, and fatty acid types were determined [43-46]. Identification was confirmed using the 16S rDNA gene which was amplified by PCR, purified and sequenced (Perkin Elmer, USA). Sequences were compared to the GenBank database as described before [47].
Oil Spreading Assay
The oil spreading test was determined by the modified method of Morikawa [48] where 100 μl of crude oil is added to the surface of 20 ml of broth medium in a Petri dish to form a thin oil layer. Then, the oil layer was mixed slowly with the supernatant, clearing zone formation mean positive results and the diameter of this zone correlates to surfactant production.
Hemolysis assay
The lysis of red blood erythrocyte cells by Biosurfactants was determined by hemolysis assay [49]. Bacteria were grown on sheep blood agar plates for 4 days at 30°C. Cell lysis means positive results.
Drop Collapse
Drop collapse test was performed with slight modifications [50, 51]. On glass slide, drop of crude oil and drop culture filtrate were added, drop collapse activity was recorded. Deionized water and Triton X-100 (1 mg/ml) were used as negative control and positive control, respectively, flat drops meaning Biosurfactant production [52].
Determination of Emulsification Activity
The emulsification index (EI24) was measured using modified method [53] where 4 ml of the biosurfactant was a vortex at 25°C with 4 ml of crude oil for 5 min. After 24 hr, the EI24 was calculated as follows: by
EI24 (%) = α / β ×100
α: the height of the emulsified layer (mm), β: total height (mm).
Reduction of surface tension
After 7 days of incubation, cultures were centrifuged at 4°C and 5000 rpm for 15 min, the surface tensionwas determined using a Tensiometer ((Model 70545, USA).
Surface tension (ST) reduction (%) = STc − STs/STc × 100
STc: ST of control, STs : ST of the sample.
Percentage of oil degradation
The percentage of oil degradation by the selected actinomycetes was determined after growth for 25 days in the MSB medium. About 0.5 g of the crude oil was added to 10 ml of the previous medium and each flask was inoculated with 0.5 ml of heavy bacterial suspension (8x107 cfu/ml). Non inoculated flasks were served as control. After the incubation period at 45°C, weight loss in the oil was determined using the following equation [54]
Weight of crude oil cotrol- weight of crude oil sample ×100weight of crude oil (cotrol)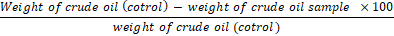
For estimation of the weight of the crude oil, 5 ml of n-hexane was added to each flask. The contents were transferred to a separating funnel and extracted. Extraction was carried out twice, the solvent was dried ad crude oil was weighted for sample and control.
Estimation of bacterial growth
Bacterial growth was measured by protein contained, measured by Bradford method [52]. Dry weight (mg/l) was determined from the standard curve of protein content and dry weight.
Biodegradation of some Hydrocarbons
Hydrocarbons (PAHs) biodegradation of phenanthrene, Fluorene, pyrene, and benzopyrene were determined in conical flasks (100 ml) containing 20 ml MSB medium and different concentrations of the tested hydrocarbon (50 to 1500 ppm) and inoculated with 2 ml of the tested bacterial suspension (8x107cfu/ml). All the flasks were incubated at 45°C at 100 rpm for 10 days and the cell growth (protein content) and biodegradation activity (HPLC analysis) were detected. Non inoculated flasks containing PAH was used as control.
The experimental flasks contain PAH were extracted twice with ethyl acetate (v/v), dried with anhydrous sodium sulphate, concentrated to 1 ml and filtered through a 0.2-mm syringe filter for quantitative analysis in Ultra-Fast Liquid Chromatography (Shimadzu). Degradation percentage was calculated from retention time and peak area compared to standard.
Statistical analysis
All the experiments were carried within triplicates and the mean value was calculated.
RESULTS
Four soil samples were collected and analyzed for organic matter and total hydrocarbon. The pH was ranged from 6.8-7.4 and EC was in the range of 0.25-0-44 µS/cm. Organic matter (%) was ranged from 2.9-5.8% while total hydrocarbon content (mg/kg) was 490, 1121, 906 and 349, respectively for the 4 tested soil samples (Table 1). These soil samples were used for actinomycetes isolation on Starch nitrate broth. About 15 actinomycetes were developed, purified and each isolate was screened in MSB medium containing 1g/l of Petroleum oil and growth was determined. The isolates MB5, MB7 MB9, MB10, and MB11 showed the highest growth in oil containing broth medium and bacterial growth and clear zone were found around their colonies. The rate of oil degradation was ranged from 50-73%. The most active isolate for oil degradation (% of oil degradation 73%) was isolated MB7 (Table 2). The five isolates were screened for biosurfactant production using different methods including drop collapse test, emulsification index, and surface tension. Particularly, the isolates MB7 MB9 and MB11 gave quick positive results for all biosurfactant screening methods, specifically for drops collapse test which indicating higher production of the biosurfactant in the culture filtrates (Table 2). In the initial screening, the Emulsification index for the tested isolates was ranged from 33-96%. For the oil spreading test, clear zones' diameter was ranged from 1.3-3.2 cm which was visualized by the addition of the culture filtrates in the crude oil layer (Fig. 1). These results confirmed the presence of biosurfactants in the cell-free culture supernatant. The most active isolate in biosurfactant production was isolate MB9 which had emulsification index 96 ± 2.4 and surface tension 29 mN/m, thus it was selected for more detail studies, characterized and identified.
The isolate MB7 formed large, rough, and irregular colonies. Growth, mycelia color and pigment production on different growth media were shown in Table 3. It was a Gram-positive, filamentous bacterium with pink color colonies. The aerial mycelia that bearing a chain of non-motile spores, produced in straight chains, with a smooth surface are found (Table 4). The isolate MB9 was examined under light and Scanning electron microscope (Fig. 2).
Different biochemical tests were carried out (Table 5), gelatinase, lipase, and protease were positive. Diagnostic sugar was not detected cells after hydrolysis while L-form of Diaminopimelic acid and, Phosphatidylethanolamine were detected using one and two dimensions thin-layer chromatography. Iso and anteiso fatty acids were detected in cell extract using Gas chromatography (Table 6). The isolated bacterium was identified based on a scheme in Bergy's Manual of Systematic Bacteriology as species belonging to genus Streptomyces. Identification was confirmed using 16SrRNA (Fig. 3).
The identified bacterium isolate MB9 was grown in minimal medium containing different concentrations of PHA as a carbon and energy source for 10 days at 45°C. After incubation at 45ᵒC, the growth was measured by measuring the protein content where there is a direct proportion between growth (dry weight) and protein content of the cell. Bacterial growth was zero for the control while increased in the treated sample which contains the bacterial consortium Percentage of degradation was measured using High-performance liquid chromatography (HPLC). It was found that growth and % of degradations were increased by increasing the incubation period. Complete degradation of 100 ppm of FLU, Phenanthrene and Naphthalene were recorded which was associated with an increase in bacterial growth while in control the degradation was negligible (Fig. 4, 5 and 6). Similarly, complete degradation for 50 ppm Anthracene and 25 ppm of Pyrene was recorded (Fig. 7 and 8). It was found that increasing PHA decreased growth and degradation rates. Out of 1500 ppm, 80% were degraded for Flu and Phenanthrene, while 30%were degraded for Naphthalene and only 10% were degraded for both Anthracene and Pyrene.
|
MB11 |
|
MB9 |
|
MB7 |
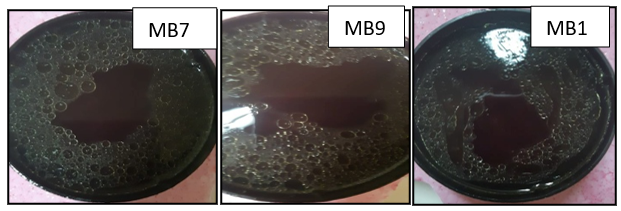
Figure 1. Oil displacement area formed by the filtrate of the isolates MB5, MB7, and MB9
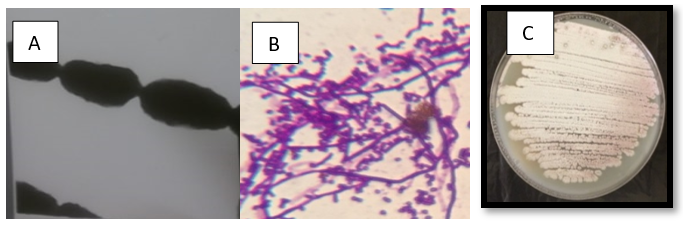
Figure 2.A: Bacterial growth on Starch nitrate agar, B: cell shape under light microscope x1000 and C: Transmission electron microscope X 30 000
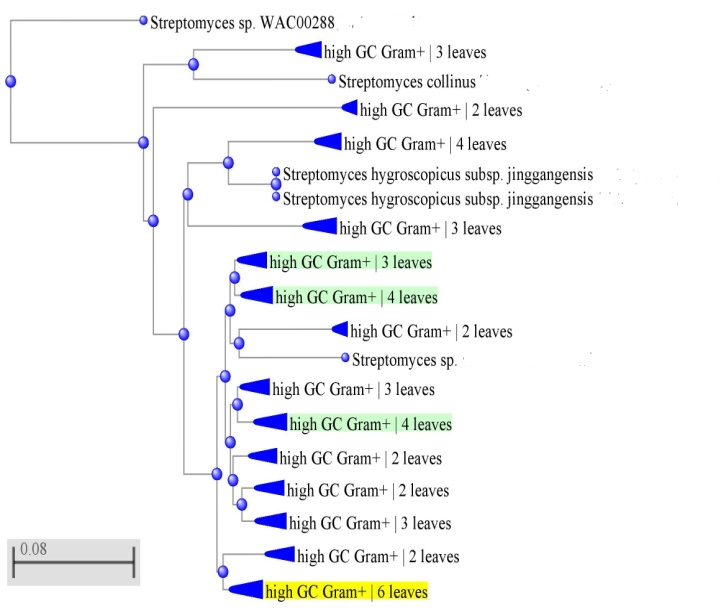
Figure 3: Phylogenetic tree of 16S rRNA of Streptomyces sp. MB9 and allied bacteria
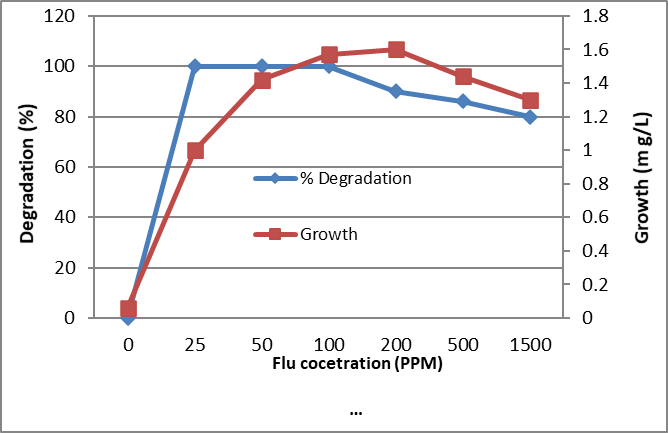
Figure 4. Biodegradation of different concentrations of Fluorene by the tested bacterium MB9 after 10 days
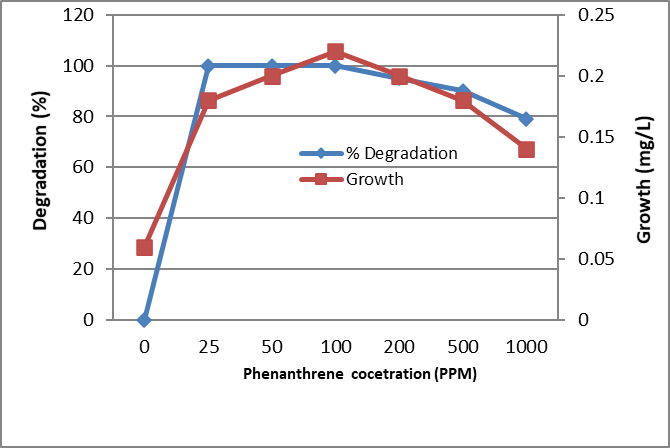
Figure 5. Biodegradation of different concentrations of Phenanthrene by the tested bacterium MB9 after 10 days
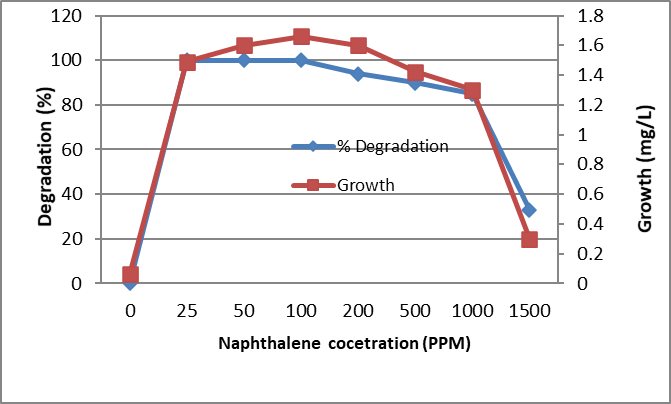
Figure 6. Biodegradation of different concentrations of Naphthalene by the tested bacterium MB9 after 10 days
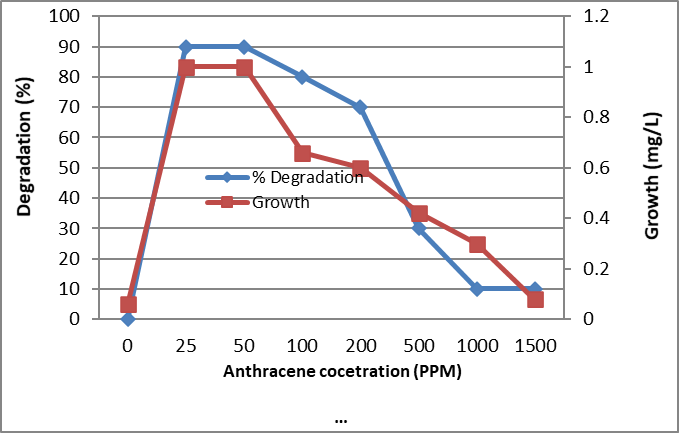
Figure 7. Biodegradation of different concentrations of Anthracene by the tested bacterium MB9 after 10 days
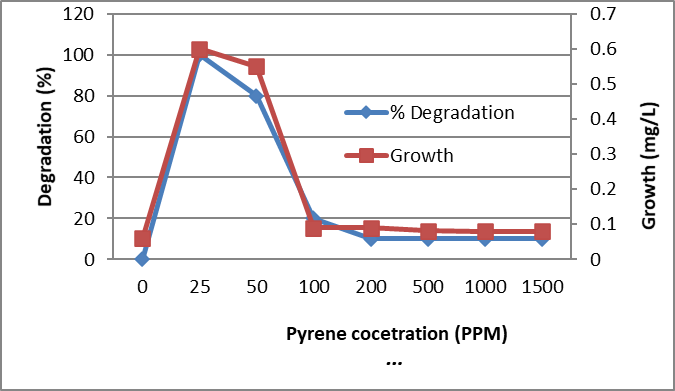
Figure 8. Biodegradation of different concentrations of Pyrene by the tested bacterium MB9 after 10 days
Table 1. Some physicochemical properties of contaminated collected soil samples
|
Soil sample (0-15 depth) |
pH |
EC (µS/cm) |
Organic matter (%) |
Total Hydrocarbon (mg/kg) |
|
Soil 1 |
7.4 |
0.29 |
2.9 |
490 |
|
Soil 2 |
7.2 |
0.25 |
4.9 |
1121 |
|
Soil 3 |
6.8 |
0.34 |
5.2 |
906 |
|
Soil 4 |
7.2 |
0.44 |
5.8 |
349 |
Table 2. The activity of the isolated actinomycetes for biosurfactant production
|
Isolate |
Color |
Hemolytic assay |
Drop collapse test |
Oil spreading assay (cm) |
Emulsification index |
Surface tension (mN/m) |
Oil degradation |
|
|
Clear zone |
% of degradation |
|||||||
|
MB5 |
Gray |
- |
- |
1.0±0.7 |
ND |
ND |
+ |
15 |
|
MB7 |
Gray |
+ |
+ |
2.8±01 |
71±11 |
40±11 |
+ |
50 |
|
MB9 |
Pink |
+ |
+ |
3.2±0.9 |
96±2.4 |
29±11 |
+ |
73 |
|
MB10 |
Black |
- |
+ |
1.0±0.0 |
40±11 |
ND |
+ |
16 |
|
MB11 |
White |
+ |
+ |
2.6±0.0 |
33±11 |
60±11 |
+ |
51 |
Table 3: Cultural characteristics of the actinomycete isolate MB9 grown on different agar medium.
|
Agar media |
Growth |
Aerial mycelium color |
Substrate mycelium color |
Soluble pigment |
|
Starch-nitrate agar |
Heavy |
Pink |
Brown |
- |
|
Glucose Asparagine |
Heavy |
Pink |
Dark yellow |
- |
|
Inorganic salts-starch iron (ISP-4) |
Moderate |
Pink |
Pale yellow |
- |
|
Tyrosine agar (ISP-7) |
Moderate |
Pale pink |
Pale yellow |
- |
|
Yeast extract-malt extract (ISP-2) |
Moderate |
Pale pink |
Pale yellow |
- |
|
Oatmeal agar (ISP-3) |
Poor |
Yellow |
Pale yellow |
- |
|
Glycerol asparagine agar (ISP-5) |
Poor |
Dark yellow |
Yellow |
- |
Table 4: Morphology of the tested isolate MB9.
|
Tested character |
Result |
|
Gram stain |
+ |
|
Source of isolation |
Oil contaminated Soil |
|
Motility of spore |
Absent |
|
Shape of spore |
Cylindrical (4-6 and 5-7 µm) |
|
Spore chain |
Straight chain |
|
Spore surface |
Smooth |
|
Number of spore/ chain |
12-18 |
|
Aerial hyphae and Substrate mycelium |
Well developed |
|
Zoospore, Sporangium, Sclerotia and Fragmented mycelia |
Absent |
Table 5: Physiological characters of the isolate MB9.
|
Character |
Result |
Utilization of Carbon or nitrogen sources |
Result |
|
Melanin pigment production on Tyrosine agar |
- |
Carbon source Glucose |
++ |
|
Chitinase |
+ve |
D-xylose |
-- |
|
Lipase |
-ve |
D-mannitol |
++ |
|
Lecithinase |
+ve |
glycerol |
++ |
|
Gelatinase |
+ve |
D- galactose |
-- |
|
Pectinase |
+ve |
Sucrose |
++ |
|
H2S production |
-ve |
Starch |
++ |
|
Growth temperature |
15 – 45ºC |
Nitrogen sources |
|
|
Tolerance to NaCl |
5-12% |
KNO3 |
++ |
|
pH range |
6-9 |
Valine |
++ |
|
Resistance to Penicillin |
+ |
Phenylalanine |
++ |
ve: negative result, +ve: positive result, +: resistance, : sensitive, ++ utilization, --: no utilization
Table 6: Chemical composition of the cell wall or cell hydrolysate of the tested isolate MB9
|
Type of the reaction |
Results |
|
- Diagnostic sugar |
Not detected |
|
- Amino acids Diaminopimelic acid (DAP) Glycine Alanine Lysine |
+ (L-form) + - - |
|
-Phospholipids Phosphatidylethanolamine (PE) Phosphatidylinositol (PI) |
+ - |
|
-Fatty Acids Iso-anteiso fatty acid |
+ |
DISCUSSION
Actinomycetes are found in different habitats and can be isolated from oil-contaminated area o starch nitrate agar [46]. All actinomycete isolates were grown in medium sublimated with 1g/l, out of 15 only 5 isolates can use petroleum oil in liquid medium as carbon source. Degradation of oil by many genera of actinomycetes especially Streptomyces, Arthrobacter, Rhodococcus, Gordonia and Mycobacterium are known hydrocarbon degraders was confirmed by may authors [55, 56] while Mereena et al. [57] reported that from actinomycetes genera, Streptomyces and Micromonospora were the most active in hydrocarbon degradation. The five isolates of actinomycetes were screened for Biosurfactant production using different methods described before [58, 59]. Many methods were developed for the rapid screening of biosurfactant producing isolates where each technique has an advantage, thus using the diverse methods is important for useful screening. Out of the 5 screened isolates, 3 decreased the surface tension and the values were similar to that found in the pieces of literature (27–42 mN/m). The presence of Biosurfactant produced by Bacillus decreased surface tension of up to 27 mN/m [59].It was reported that bacteria producing Biosurfactants were distributed in different habitats especially in oil-contaminated soil which yield more oil degraded bacteria compared to uncontaminated soils soil [49]. The kerosene fungus, Cladosporium resinae was isolated from aircraft fuel tank [58]. Biosurfactants were obtained from organisms that were isolated from oil-contaminated soils [60]. In various industrial fields, the most important surfactants were used for emulsification, corrosion inhabitation, and detergent production. Although surfactant production was costly, it was increased every day, thus the production of biosurfactants through fermentation is studied [59].
The most active isolate for Biosurfactant production was isolated MB9 which was characterized and identified as species belonging to genus Streptomyces. Identification was confirmed using 16SrRNA and it was identified as Streptomyces sp. MB9. The identified Streptomyces sp. MB9 was used for PAHs degradation in liquid medium and growth and % of degradation were measured after 10 days of growth. The degradation process was decreased by increasing the concentration of the tested material. It is clear that actinomycetes especially genus Streptomyces played an interesting role in the removal of crude oil from soil [61]. Similarly, Streptomyces sp. ERI-CPDA-1 was isolated from contaminated soil and showed excellent activity for polycyclic aromatic hydrocarbons degradation where 98.25% of diesel oil, 99.14% of naphthalene and 17.5% for phenanthrene were removed in 7 days at 30°C. The endophytic actinobacteria to degrade crude petroleum, Six of Streptomyces genus, out of 17 isolates, were tolerant to petroleum on solid minimal medium and five strains could use petroleum as sole carbon and energy source, 98% removal after 7 days of incubation. They added that these isolates produced biosurfactants and used n-alkanes or polycyclic aromatic hydrocarbons [62]. In Conclusion, petroleum products contained products that can enhance bacterial growth or are not available to some bacteria. The presence of these highly toxic petroleum hydrocarbons with recalcitrant properties led to environmental contamination.
ACKNOWLEDGEMENTS
This Project was funded by the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, under grant No. (J-412- 247- 38). The author, therefore, acknowledges with thanks to DSR technical and financial support. The author also thanks Dr: Magda M. Aly, Prof of Microbiology, Faculty of Science, KAU, for help and revising the manuscript.
REFERENCES